Having trouble figuring out what a fish has eaten? Just use DNA!

Figure 1. A smallmouth bass captured swimming underwater. Photo: Wikimedia Commons
Ever wonder how scientists figure out what fish eat – or why they are even curious about fish diets? One answer to these questions comes from a popular sport fishery in Lake Ontario for smallmouth bass (Micropterus dolomieu; Figure 1) and largemouth bass (Micropterus salmoides; Figure 2). Typically, scientists remove or flush a stomach and examine the gut contents using a magnifying lens or scope. Bass are piscivorous (meaning they eat other fish) and can digest prey quickly. Imagine if you had to identify a fish if it was just a chunk of muscle and a couple bones!

Figure 2. A largemouth bass captured swimming underwater. Largemouth bass can be differentiated from smallmouth bass by the dark horizontal line across their body and a mouth that extends past their eyes. Photo: Bemep, Flickr.
Unsurprisingly, the rates of successful identification are slow and low. However, a new method for assessing stomach contents was borrowed from the field of human medicine and has proven to be powerful and reliable: DNA sequencing. Erich Nelson and his colleagues from the Freshwater Fisheries Conservation Lab at Queen’s University and the Lake Ontario Management Unit tested the efficacy of DNA barcode sequencing for prey identification in smallmouth and largemouth bass diets. Sport fishery managers generally want stable populations of big, healthy fish. Reliable diet data is valuable for learning which prey the fish need for growth and for tracking potential sources of contaminants in the populations.
The researchers visually analyzed the stomach contents of adult smallmouth and largemouth bass, then saved and froze the contents for DNA barcode analysis. Luckily, DNA analysis works on partially digested material, as long as some soft tissue (such as fat or muscle) remains. The researchers knew from prior research that bass primarily feed on crayfish and small fish, so they obtained primers for each of the common prey species. Primers are used to identify DNA sequences that belong to a specific species, similar to scanning a barcode at your local supermarket to show what product you have and the associated price (Figure 3). DNA was extracted from the frozen stomach content samples and combined with each common prey primer. If the primer exists in the sample, it will inform the researchers, similar to the way a barcode sticker can help grocers distinguish between different types of oranges that may visually look alike.
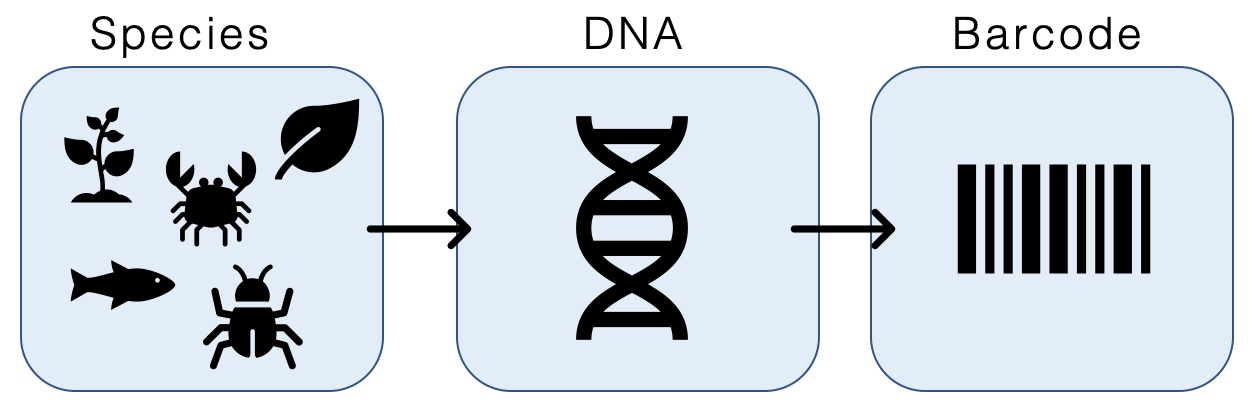
Figure 3. DNA is collected for various species. A barcode is then made specific to a portion of a species DNA that is consistent among individuals. Sample tissues can then be ‘scanned’ with this barcode, and if the species is present, the barcode will appear. Photo: Wikimedia Commons
DNA barcoding proved to be a success. This technique led to dramatically higher identification rates of stomach contents compared to visual analysis alone. In smallmouth bass, an amazing 97% of the stomach contents could be identified using DNA barcoding analysis, compared to the mere 22% that could be visually identified. Smallmouth bass diets majorly consisted of round goby (Neogobius melanostomus; Figure 4), an invasive fish species, with a small amount of alewife (Alosa pseudoharengus). DNA barcoding identification rates for largemouth bass stomach contents were 82%, compared to the 10% visually identified. The round goby was also the most consumed prey for largemouth bass, however largemouth bass appeared to have more diverse diets, also relying on bluegill sunfish (Lepomis macrochirus) and alewife for sustenance.

Figure 4. A round goby held in an experimental laboratory tank at McMaster University. Photo: Aquatic Behavioural Ecology Lab (ABEL).
These results show how an invasive species can impact local food webs and predator–prey dynamics. In this case, smallmouth and largemouth bass are growing larger by eating round goby. While big bass make for happy anglers, their growth may come with a cost. Round goby often hold high levels of contaminants in their tissues, which can accumulate in the tissues of predators like bass and render them unsafe for consumption by humans. Bioaccumulation has also been linked to lower reproductive success and unhealthy offspring in other species Without DNA sequencing and barcoding, fisheries managers might not have realized just how many round goby these bass were eating, and that the health of the populations could be at risk. Studies like this show how technological advancements such as DNA techniques can be a useful tool not just for humans, but in the world of aquatic ecology as well.
Reference:
Nelson, E. J. H, Holden, J., Eves, R., and Tufts, B. 2017. Comparison of diets for Largemouth and Smallmouth Bass in Eastern Lake Ontario using DNA barcoding and stable isotope analysis. PLoS ONE 12(8): e0181914. https://doi.org/10.1371/journal.pone.0181914



