Baited video cameras and environmental DNA create better picture of fish community
Fish can be tricky to count. Many fish are sensitive to disturbances in their surrounding environment, such as from an approaching diver conducting surveys, so care must be taken to ensure that survey methods accurately account for the fish that are present. Understanding the number and diversity of fish species within a specific habitat is important for managers and conservationists to be able to determine whether an ecosystem is healthy.
One approach for assessing fish abundance is baited remote underwater video systems (BRUVs). These systems are set up with a video recorder opposite a small container of bait that attracts fish from nearby habitat. The video recording is then analyzed to determine the species and biomass of fish present within a specific location. This method is an exceptional non-invasive approach for gaining insight into the fish community present at a particular site. However, BRUVs are not well suited to capture cryptic fish species or fish that are less mobile. BRUVs are also limited to a specific field of view and can miss species that prefer habitat not included within view of the video recorder.
Another non-invasive method for assessing fish diversity, environmental DNA (eDNA), is not reliant on visuals to determine what species are present. Instead, eDNA uses genetic material left behind when a fish swims through the water (sources include blood and excrement) to identify which species are using the habitat. Seawater is sampled, filtered and the DNA left behind analyzed and matched to known genetic sequences of fish species.
A recent study released in Conservation Biology demonstrated the value of combining these two approaches to generate a more accurate and effective assessment of fish abundance. Michael Stat at Macquarie University in Sydney, Australia and colleagues proposed that using BRUVs in conjunction with eDNA sampling can positively influence our ability to accurately evaluate and monitor fish biodiversity.
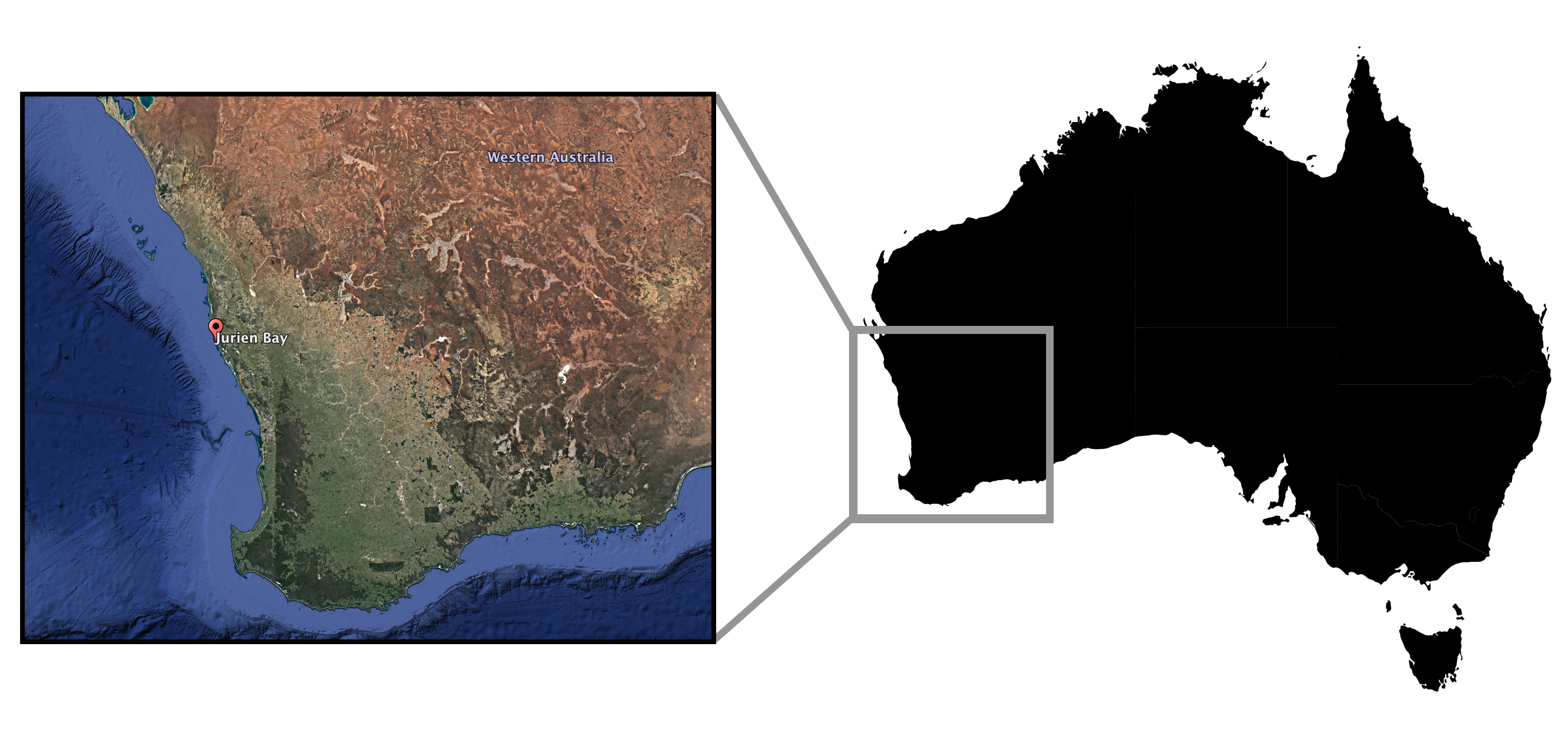 Stat and colleagues situated their field study in Jurien Bay (Western Australia) and included sites in both reef and seagrass habitats. At 48 sites, they first collected seawater samples to be used for eDNA analysis and then deployed the BRUVs for a recording period of one hour. The sample locales were an equal mix of rocky reefs (n=24) and seagrass beds (n=24) located both within and adjacent to a marine reserve.
Stat and colleagues situated their field study in Jurien Bay (Western Australia) and included sites in both reef and seagrass habitats. At 48 sites, they first collected seawater samples to be used for eDNA analysis and then deployed the BRUVs for a recording period of one hour. The sample locales were an equal mix of rocky reefs (n=24) and seagrass beds (n=24) located both within and adjacent to a marine reserve.
Both methods showed a significant difference in fish community composition between the two habitat types. The use of these two methods together increased the observed generic richness by 30% compared to what could be observed by either method independently. While the BRUVs method identified more fish species that were present in both the reef and seagrass sites, the eDNA method was useful for detecting smaller fish species that are not captured in the video recordings because they are cryptic or have nocturnal habits. Interestingly, eDNA did not detect larger predator species in this study. Stat and colleagues suggest they may have missed large predators because they are only present in relatively low individual numbers at Jurien Bay, and may not have left enough genetic material to be detected. However, they would have been attracted to the bait bag and recorded using BRUVs.
The results of this study demonstrate the value of pairing two non-invasive sampling methods in order to produce a better understanding of fish biodiversity within an area. Each method provides information that the other does not capture effectively while also producing overlap for comparison. The eDNA method is better for capturing small, cryptic species and the BRUVs may capture larger fish that are present in low numbers. When used together, these methods result in a more accurate characterization of the diversity of fish communities in marine ecosystems.
Citation: Stat, M., John, J., DiBattista, J.D., Newman, S.J., Bunce, M., Harvey, E.S. (2018). Combined use of eDNA metabarcoding and video surveillance for the assessment of fish biodiversity. Conservation Biology, doi: 10.1111/cobi.13183.
Thank you to Dr. Michael Stat for feedback on this post.